Methylator, a DNA methylation analysis workflow
Maintained by BiBs. Last update : 25/04/2024. Methylator v0.1.
Methylator is a complete Snakemake workflow to analyse DNA methylation data. Methylator runs in a dedicated Apptainer image to allow for reproducibility and was optimized to compute effciently the data on HPC clusters. We aim to make those complex analyses do able by biologists with no or little bioinformatics background. Implemented by BiBs-EDC, this pipeline runs effectively on both IFB and iPOP-UP clusters. If you encounter troubles or need additional tools or features, you can create an issue on the GitHub repository or email directly BiBs.
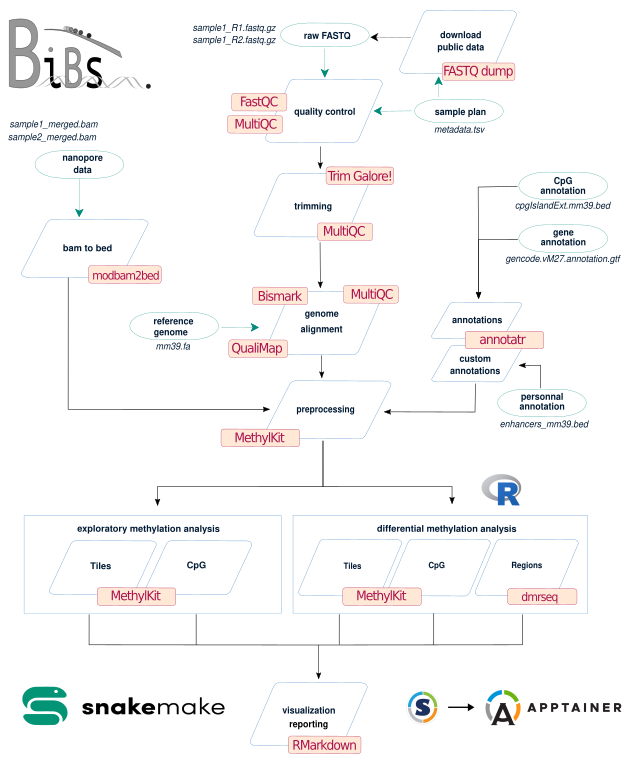
Pipeline scheme
Warning
This is a BETA version of the workflow, and of this documentation ! There is no guarantee !
If you use this workflow to analyse your data, don't forget to acknowledge BiBs in all your communications !
Acknowledge BiBs example
For EDC people : We thank the Bioinformatics and Biostatistics Core Facility, Paris Epigenetics and Cell Fate Center for bioinformatics support.
For External users : We thank the Bioinformatics and Biostatistics Core Facility, Paris Epigenetics and Cell Fate Center for sharing their analysis workflows.